|
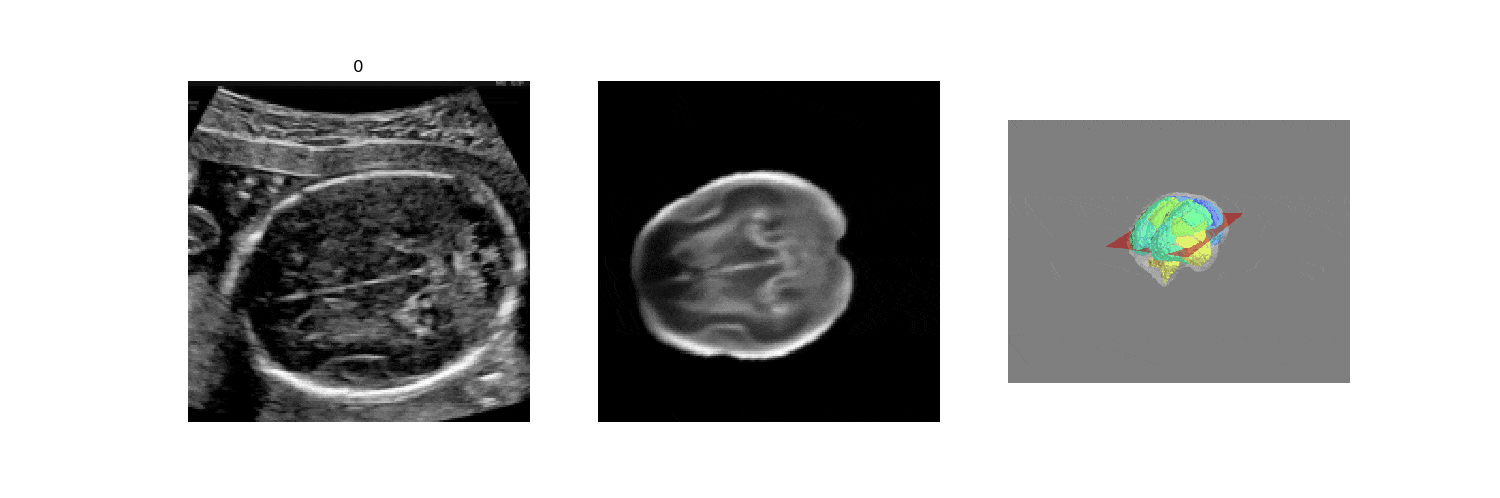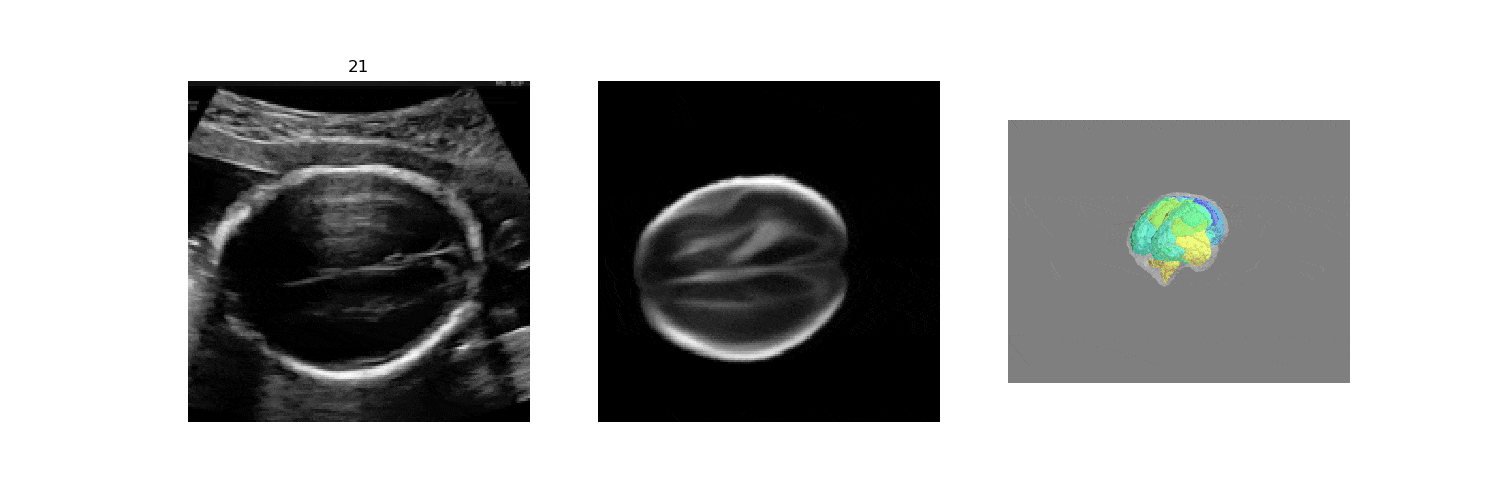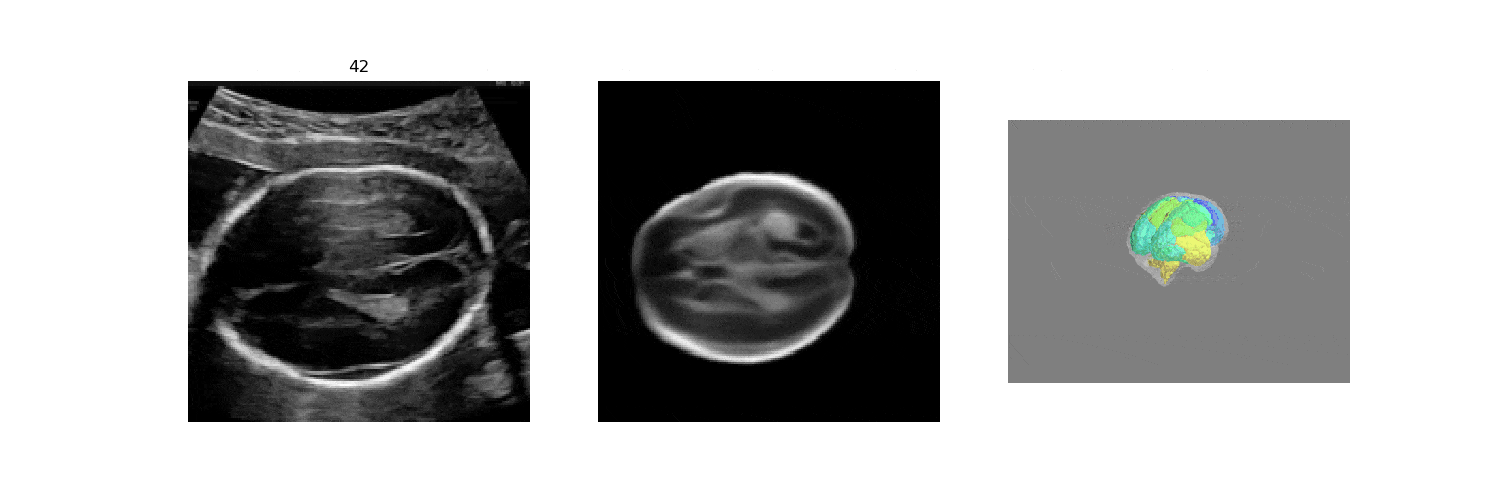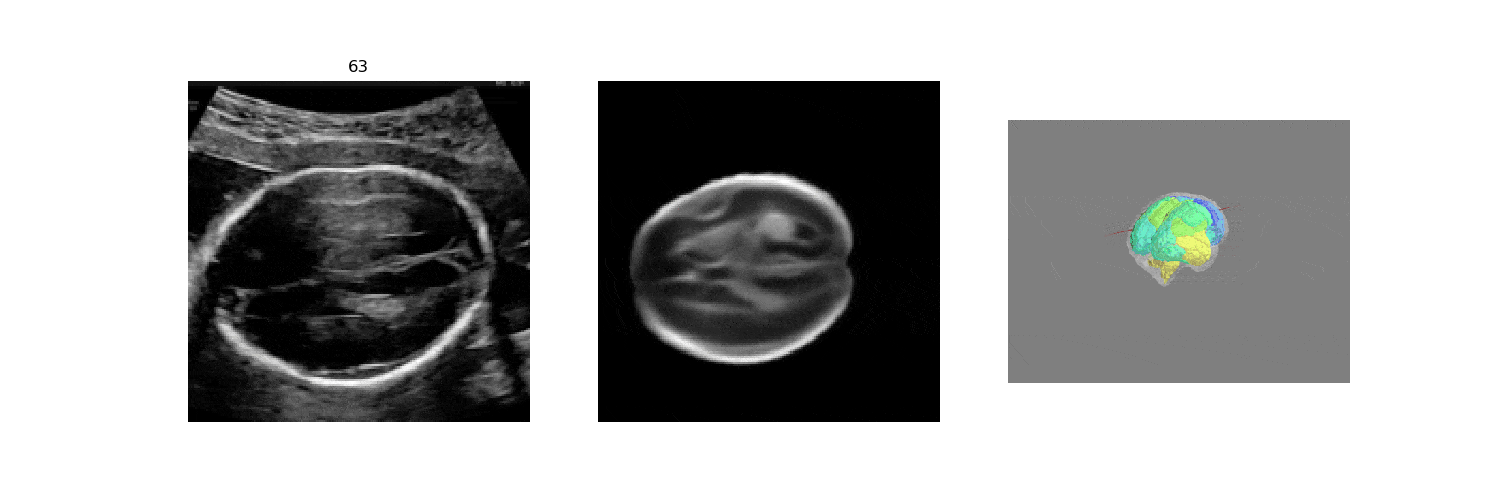
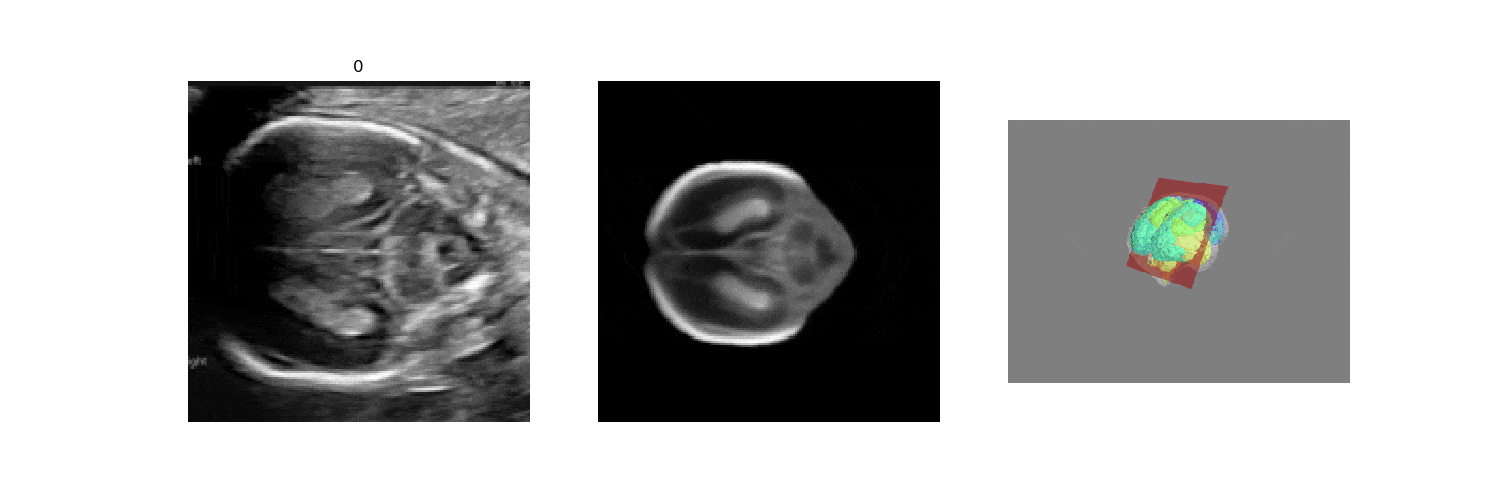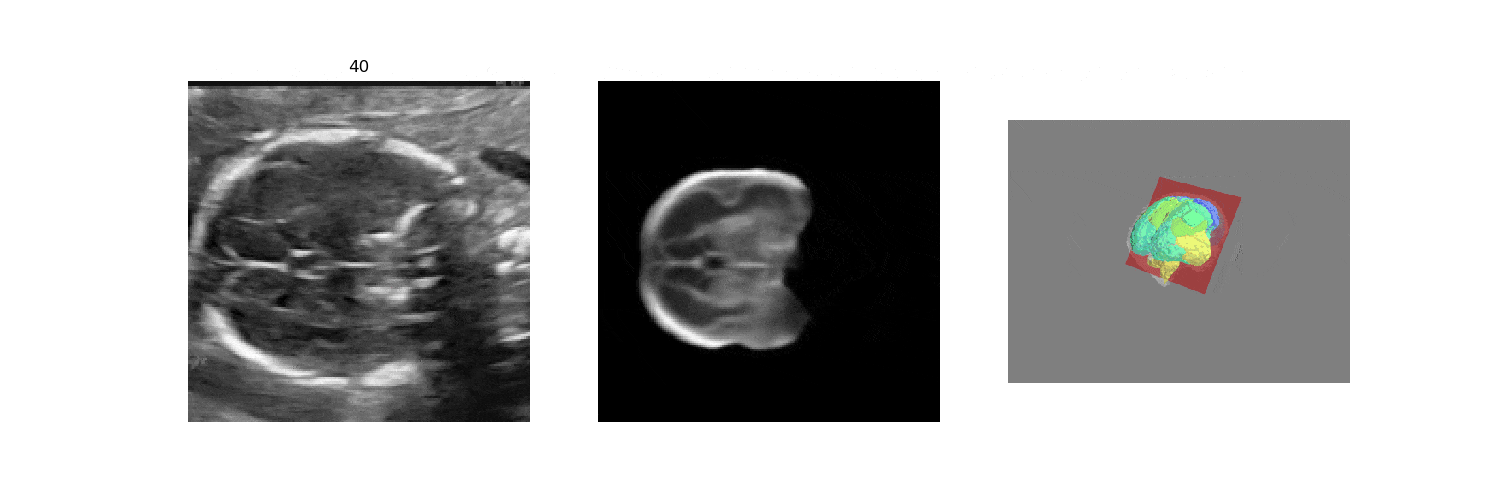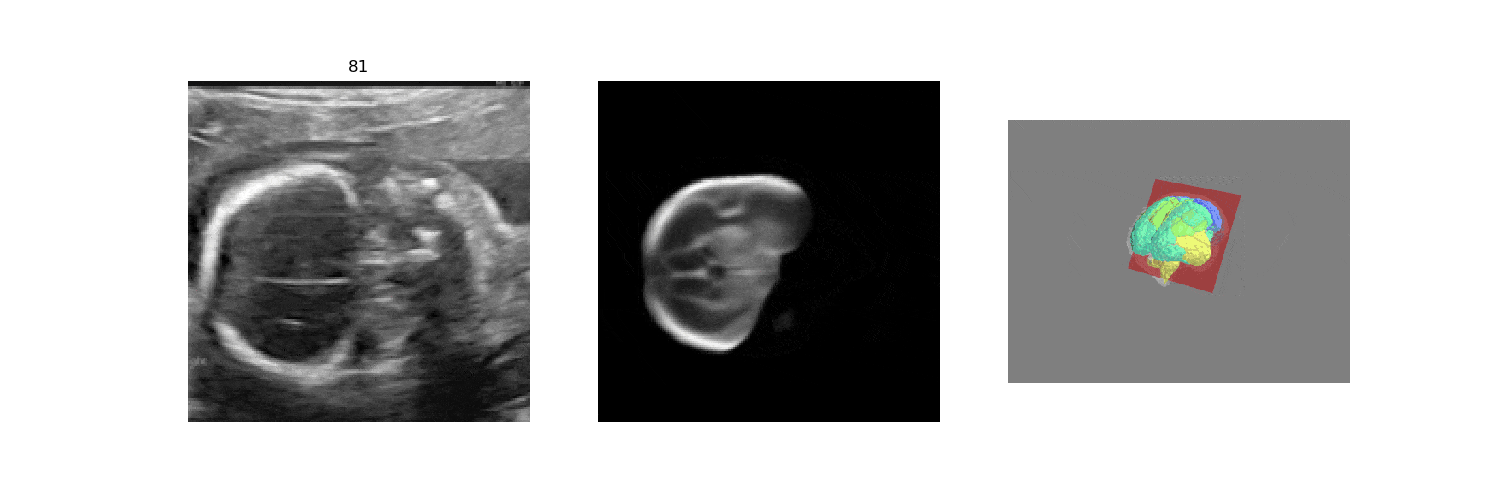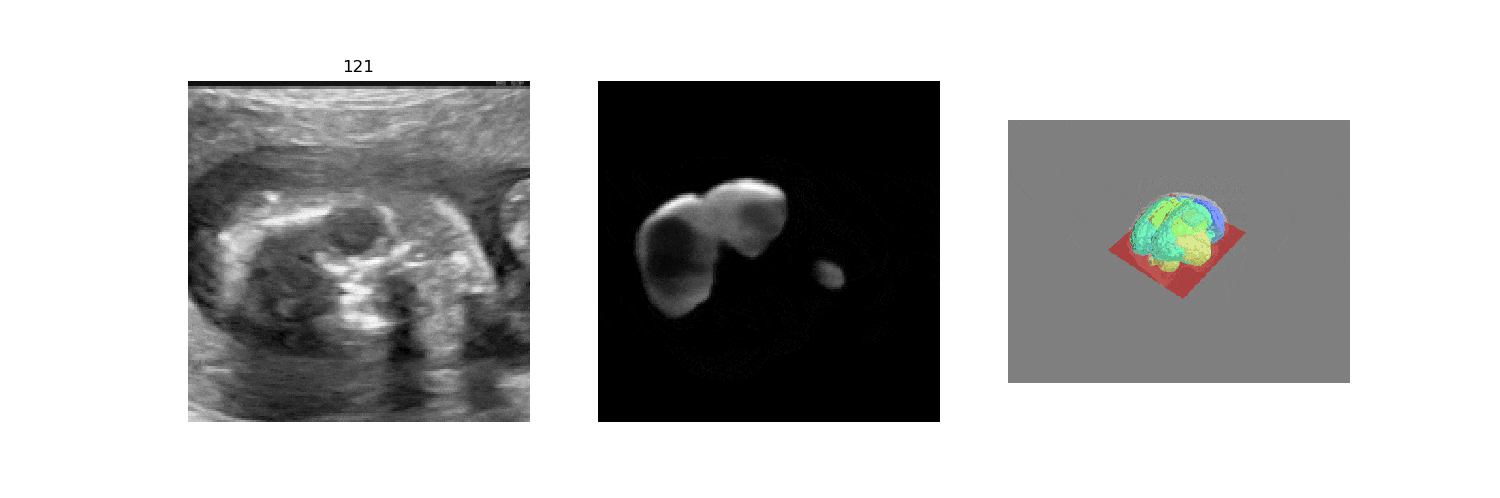
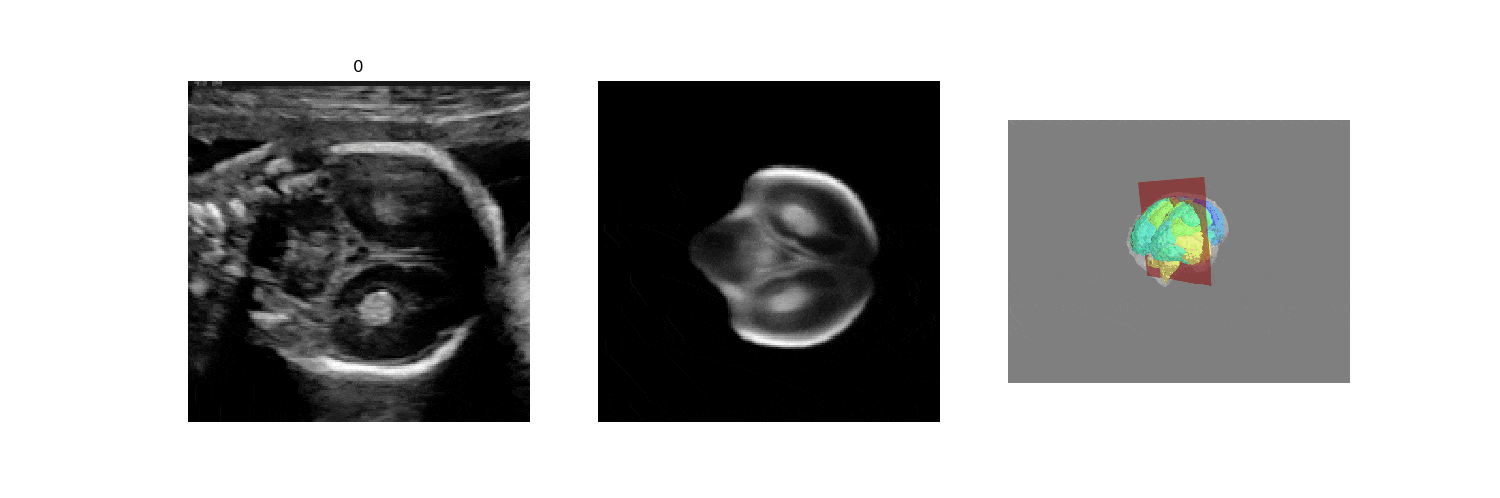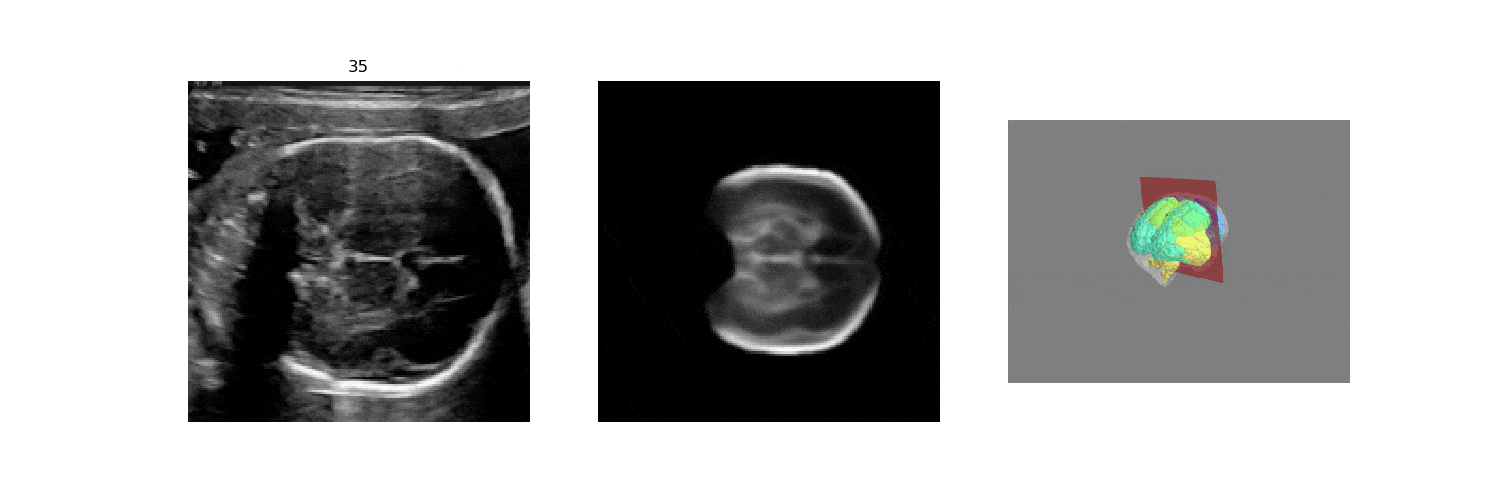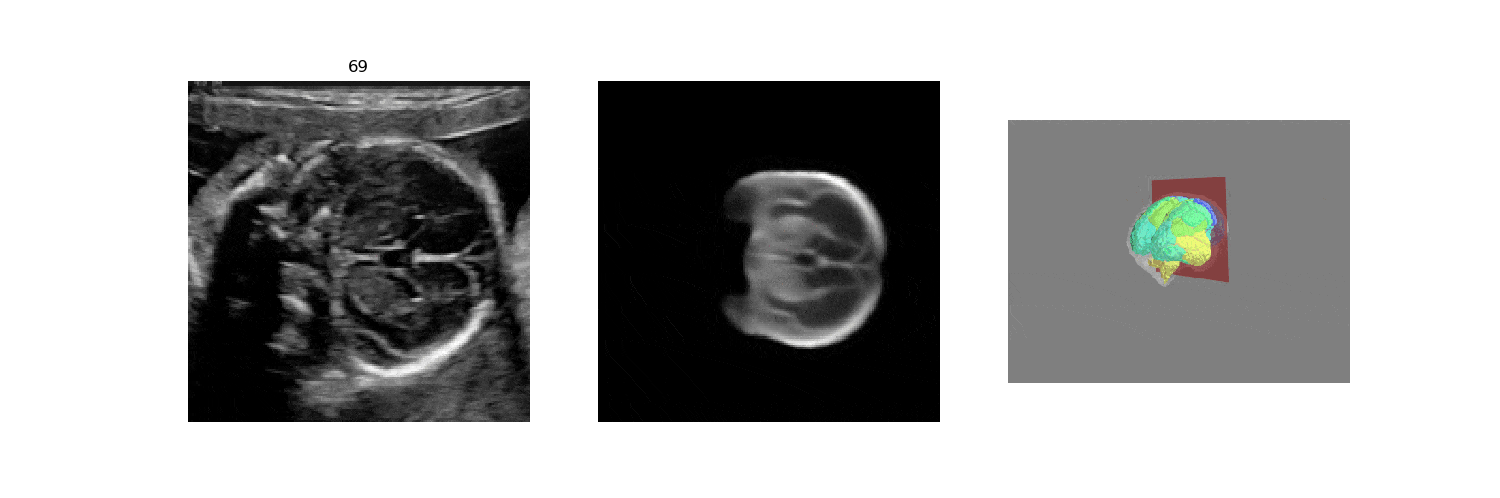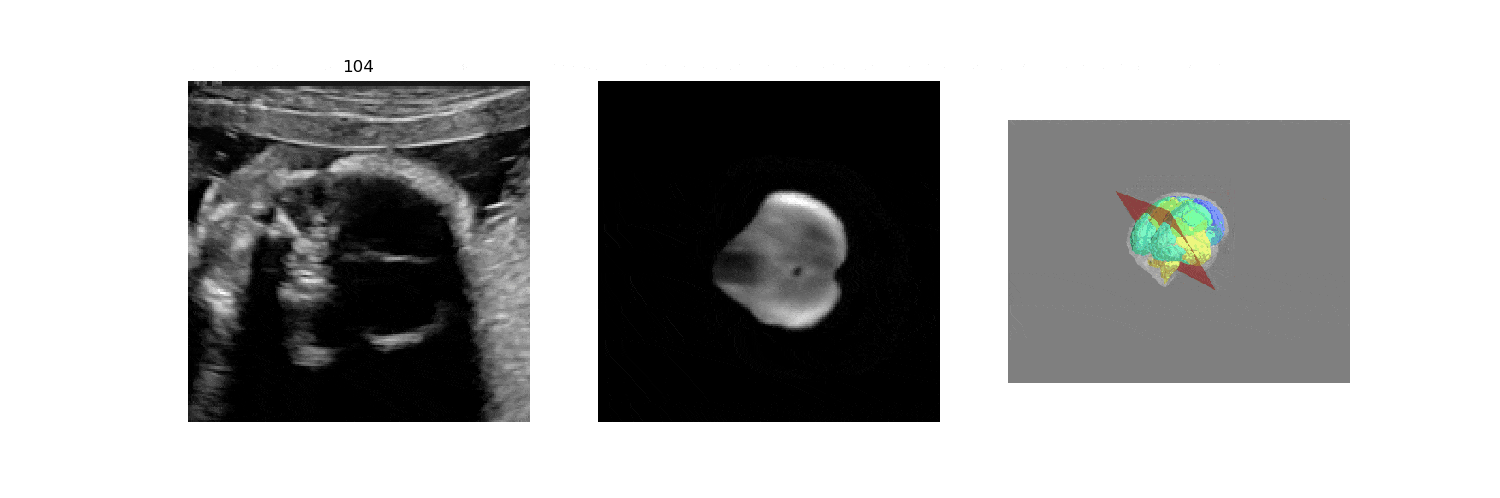
In fetal neurosonography, aligning two-dimensional (2D) ultrasound scans to their corresponding plane in the three-dimensional (3D) space remains a challenging task. In this paper, we propose a convolutional neural network that predicts the position of 2D ultrasound fetal brain scans in 3D atlas space. Instead of purely supervised learning that requires heavy annotations for each 2D scan, we train the model by sampling 2D slices from 3D fetal brain volumes, and target the model to predict the inverse of the sampling process, resembling the idea of self-supervised learning.
We propose a model that takes a set of images as input, and learns to compare them in pairs. The pairwise comparison is weighted by the attention module based on its contribution to the prediction, which is learnt implicitly during training. The feature representation for each image is thus computed by incorporating the relative position information to all the other images in the set, and is later used for the final prediction.
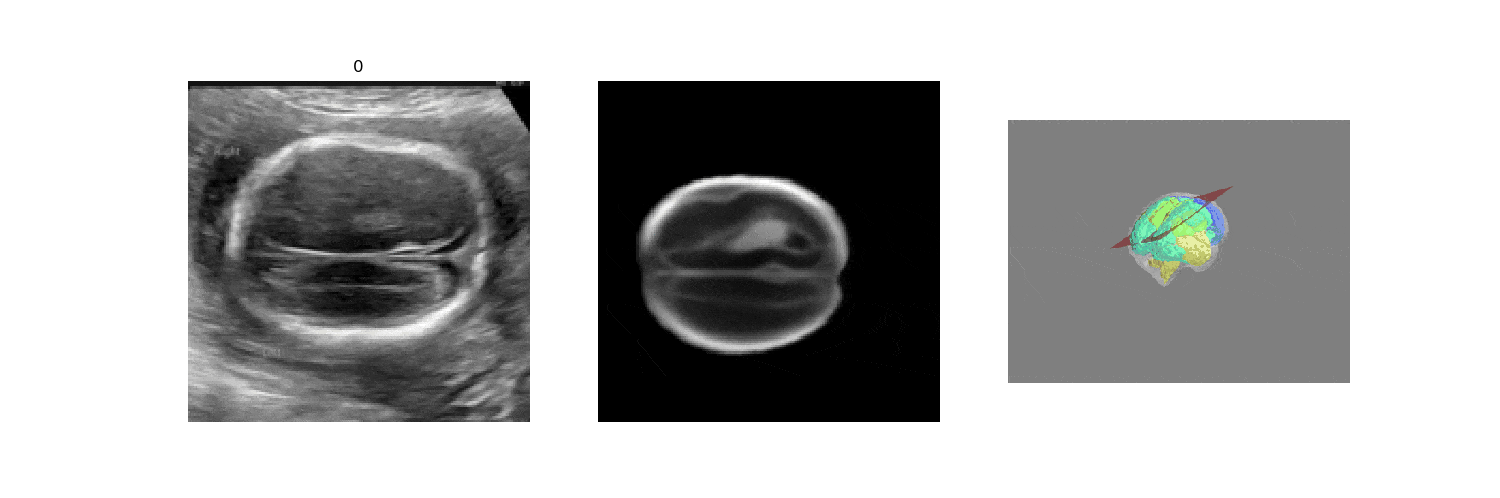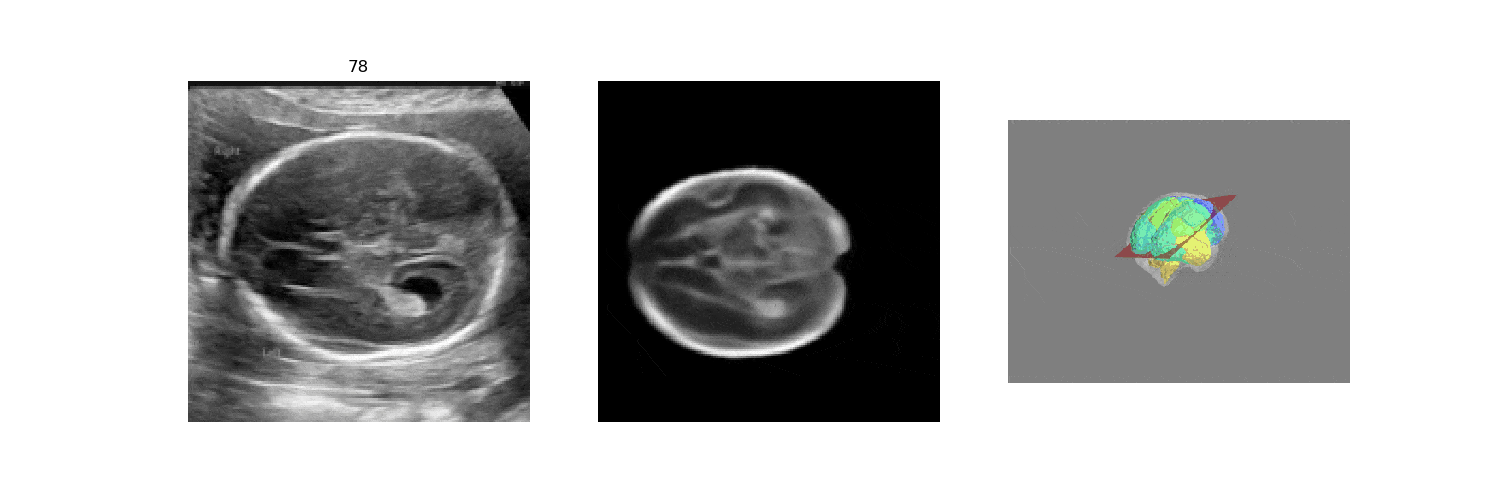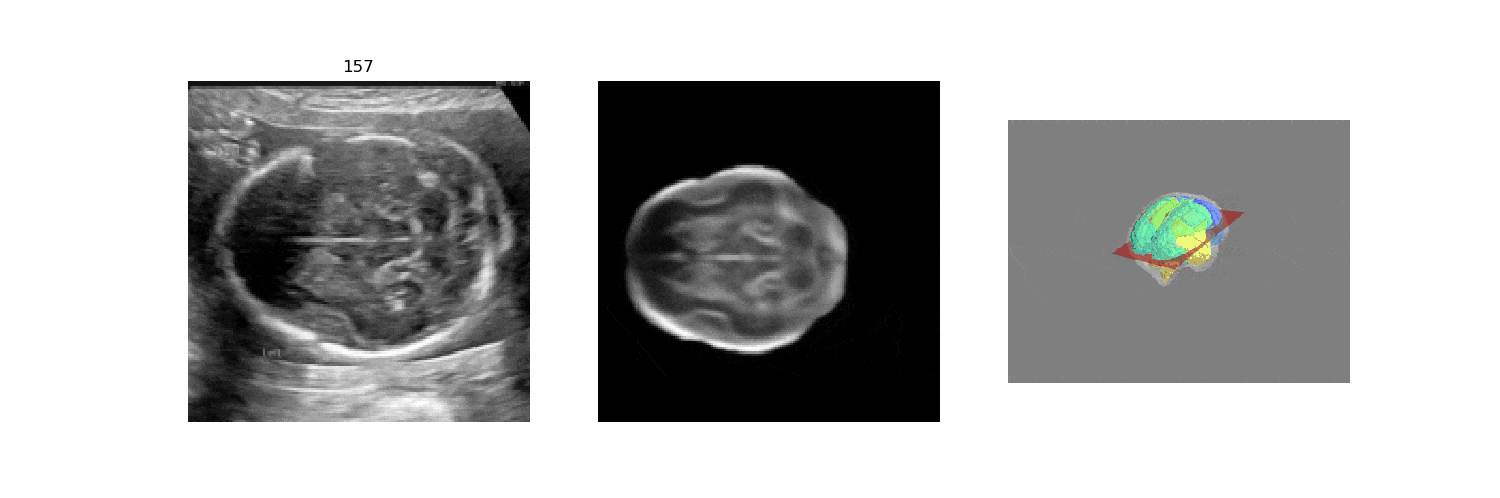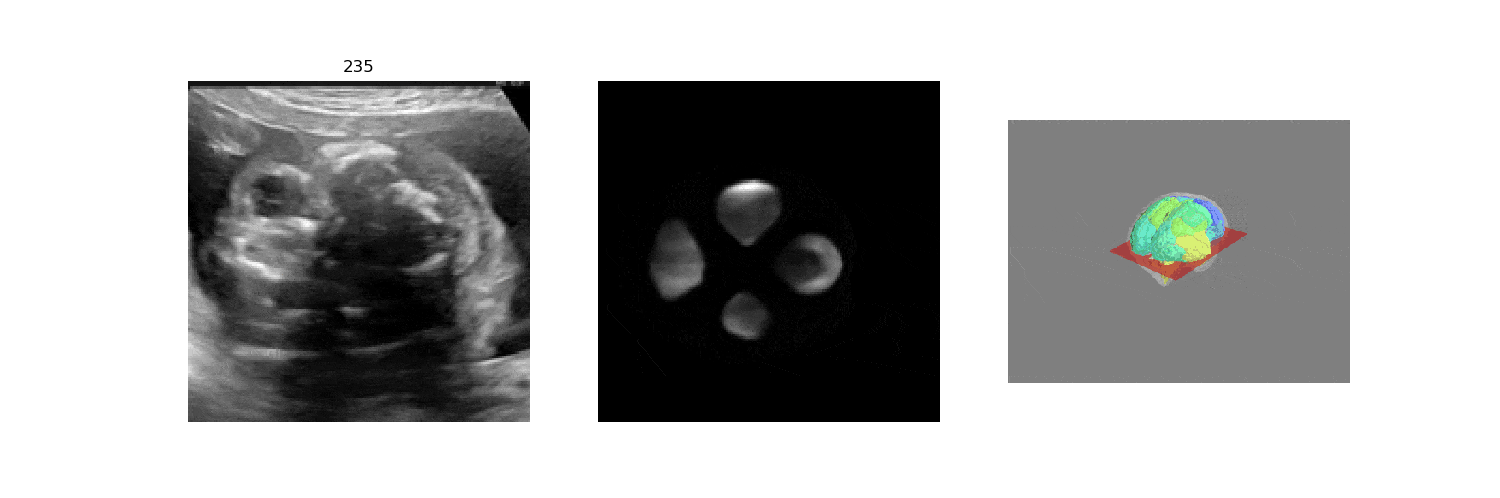
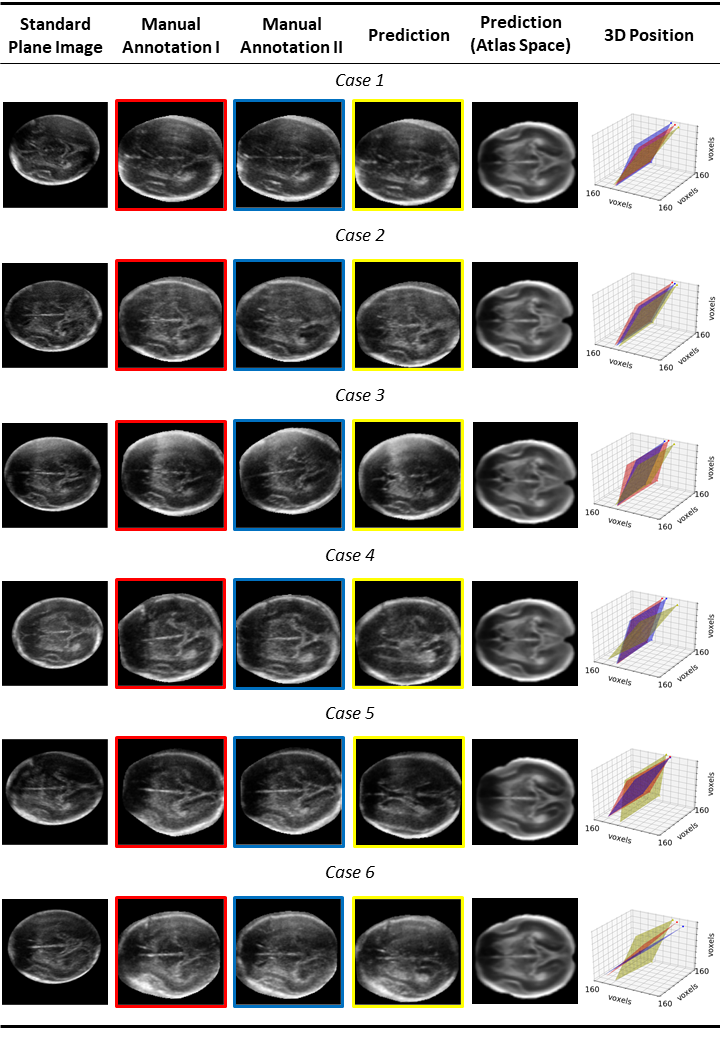
We benchmark our model on 2D slices sampled from 3D fetal brain volumes at 18 to 22 weeks' gestational age. Using three evaluation metrics, namely, Euclidean distance, plane angles and normalized cross correlation, which account for both the geometric and appearance discrepancy between the ground-truth and prediction, in all these metrics, our model outperforms a baseline model by as much as 23%, when the number of input images increases. We further demonstrate that our model generalizes to (i) real 2D standard transthalamic plane images, achieving comparable performance as human annotations, as well as (ii) video sequences of 2D freehand fetal brain scans.
|