
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|

|
|
|
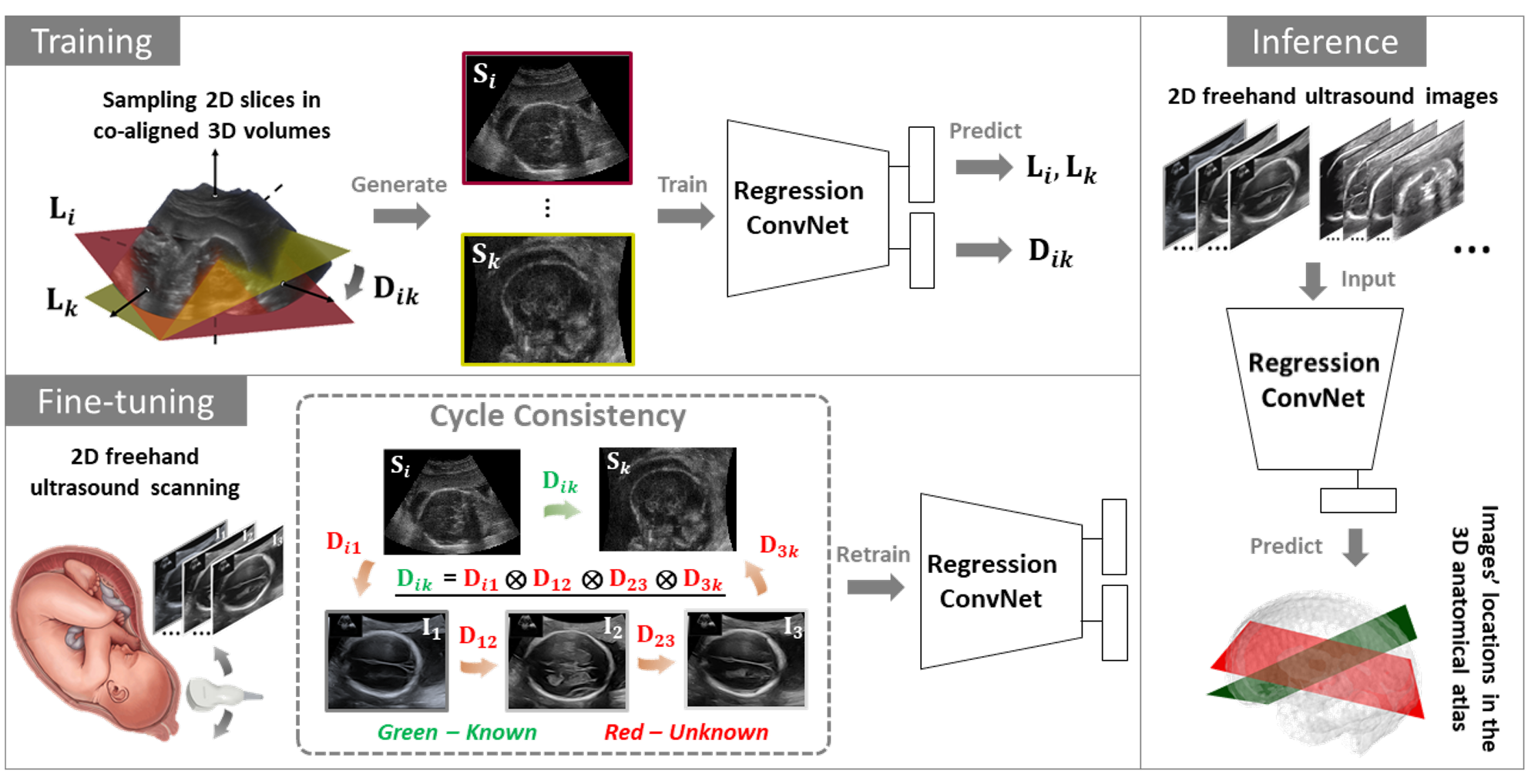
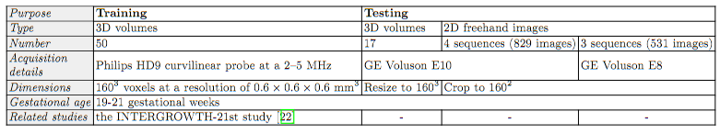
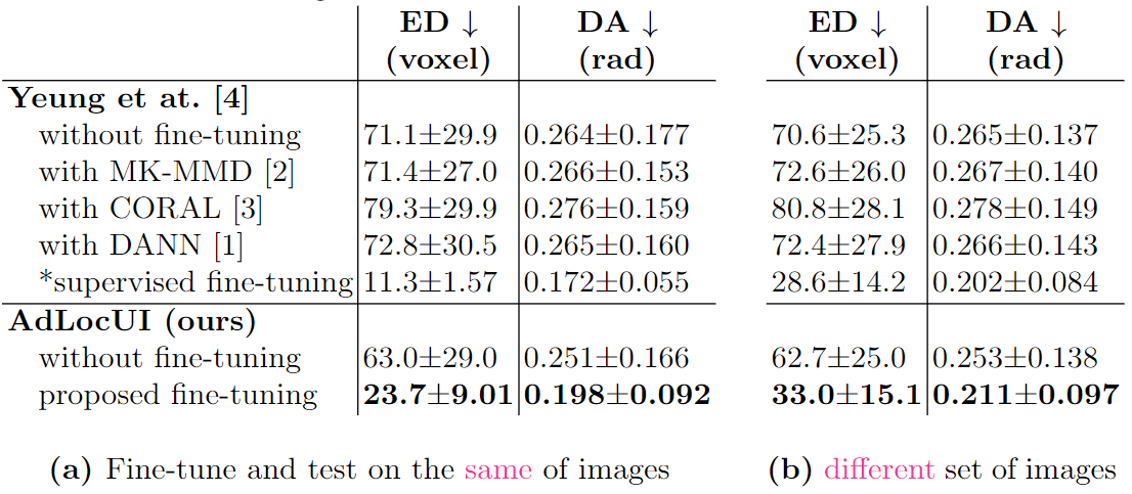
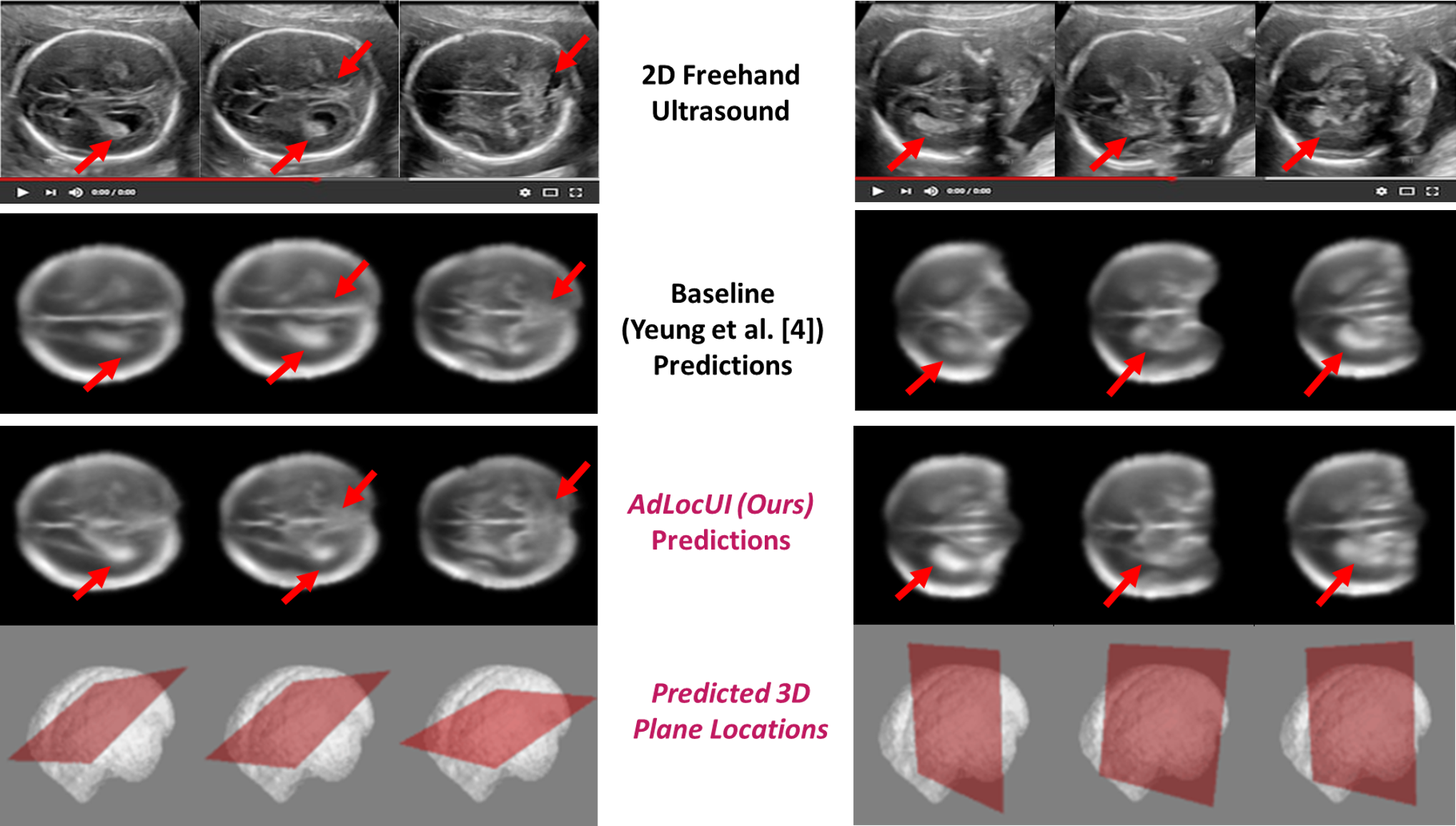
| Two-dimensional (2D) freehand ultrasound is the mainstay in prenatal care and fetal growth monitoring. The task of matching corresponding cross-sectional planes in the 3D anatomy for a given 2D ultrasound brain scan is essential in freehand scanning, but challenging. We propose AdLocUI, a framework that Adaptively Localizes 2D Ultrasound Images in the 3D anatomical atlas without using any external tracking sensor. We first train a convolutional neural network with 2D slices sampled from co-aligned 3D ultrasound volumes to predict their locations in the 3D anatomical atlas. Next, we fine-tune it with 2D freehand ultrasound images using a novel unsupervised cycle consistency, which utilizes the fact that the overall displacement of a sequence of images in the 3D anatomical atlas is equal to the displacement from the first image to the last in that sequence. We demonstrate that AdLocUI can adapt to three different ultrasound datasets, acquired with different machines and protocols, and achieves significantly better localization accuracy than the baselines. AdLocUI can be used for sensorless 2D freehand ultrasound guidance by the bedside. |
|
|
|
|
Bibtex
@inproceedings{yeung2022adaptive,
title={Adaptive 3D Localization of 2D Freehand Ultrasound Brain Images},
author={Yeung, Pak-Hei and Aliasi, Moska and Haak, Monique and the INTERGROWTH-21ST Consortium
and Namburete, Ana IL and Xie, Weidi},
booktitle={International conference on Medical Image Computing and Computer Assisted Intervention},
pages={},
year={2022}
}
|
AcknowledgementsThis template of this project webpage was originally made by Phillip Isola and Richard Zhang for a colorful ECCV project; the code can be found here. |